Abstract
Parkinson's disease (PD) is the second most prevalent neurodegenerative disorder characterized by the loss of dopaminergic neurons in the substantia nigra (SN) of the brain. Despite decades of studies, the precise pathogenic mechanism of PD is still elusive. An unbiased proteomic analysis of PD patient’s brain allows the identification of critical proteins and molecular pathways that lead to dopamine cell death and α-synuclein deposition and the resulting devastating clinical symptoms. In this study, we conducted an in-depth proteome analysis of human SN tissues from 15 PD patients and 15 healthy control (HC) individuals combining Orbitrap mass spectrometry with the isobaric tandem mass tag (TMT)-based multiplexing technology. We identified 10,040 proteins with 1,140 differentially expressed proteins in the SN of PD patients. Pathway analysis showed that the ribosome pathway was the most enriched one, followed by GABAergic synapse, retrograde endocannabinoid signaling, cell adhesion molecules (CAMs), morphine addiction, Prion disease, and Parkinson's disease pathways. Strikingly, the majority of the proteins enriched in the ribosome pathway were mitochondrial ribosomal proteins (mitoribosomes; MRPs). The subsequent protein-protein interaction (PPI) analysis and the weighted gene co-expression network analysis (WGCNA) confirmed that the mitoribosome is the most enriched protein cluster. Furthermore, the mitoribosome was also identified in our analysis of a replication set of 10 PD and 9 HC SN tissues. This study provides potential disease pathways involved in PD and paves the way to study further the pathogenic mechanism of PD.
Methods
Human SN tissues from 15 PD patients and 15 HC individuals that were used for the acquisition of the discovery data and the human SN tissues from 10 PD patients and 9 HC individuals that were used for the acquisition of the replication data were acquired from the Brain Resource Center at Johns Hopkins University School of Medicine. The clinical information for the samples is provided in Table 1 and Supplemental Table S1. Diagnosis of PD was based on UK Brain Bank clinical criteria and then autopsy confirmation (32, 33). Healthy controls were individuals without clinical or neuropathological evidence of Parkinsonism. All participants agreed to autopsy prior to their death and their next of kin consented to the autopsy procedure at the time of death. All research was approved by the Johns Hopkins Institutional Review Board. The inclusion criteria for PD are patients with 1) a clinical history of PD with or without dementia; 2) neuropathology changes of Lewy body disease brainstem-predominant, limbic, or neocortical (34); and 3) age > 50 years, males and females and any race. The exclusion criteria for PD are patients with any significant neurodegenerative or vascular comorbidity. This study abided by the Declaration of Helsinki principles.
Results
- Quantitative proteome analysis of SN samples
- Gene set enrichment analysis
- Interactome analysis
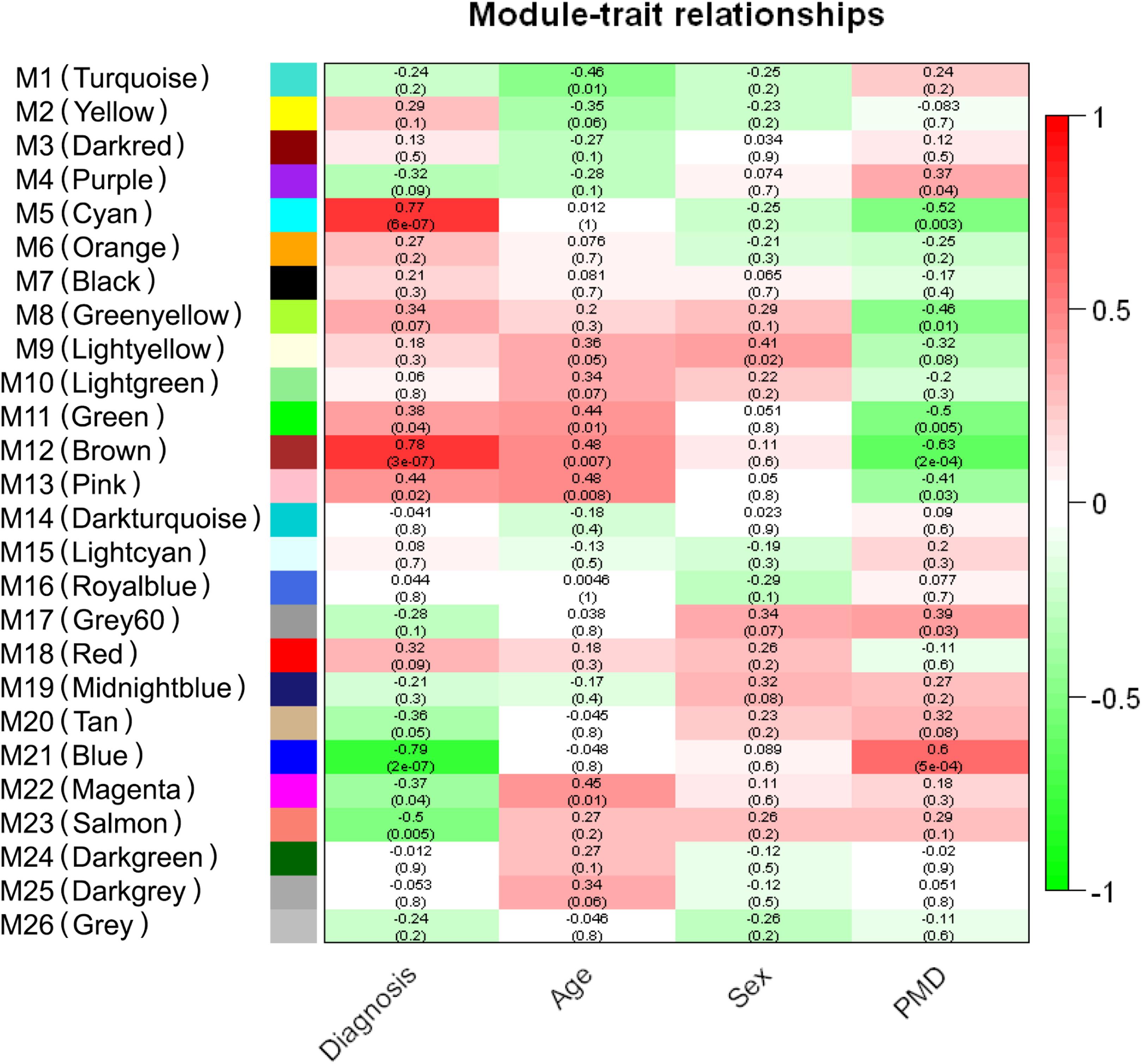
- Coexpression analysis using WGCNA
- An independent replication experiment of the pathways discovered in the main experiment
View Full Article: https://www.mcponline.org/article/S1535-9476(22)00260-2/fulltext